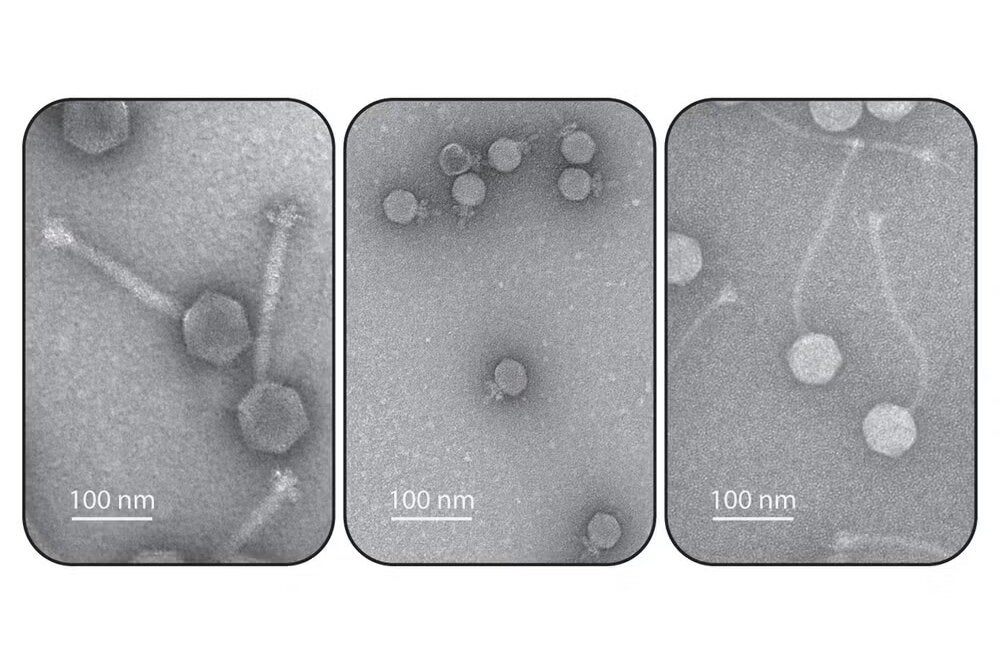
Researchers from the University of Illinois have reported complementary discoveries that will improve scientists’ understanding of immune systems in bacteria and equip clinicians with more effective options for treating antibiotic-resistant infections.
Antibiotic-resistant bacteria known as superbugs are dangerous because they are difficult to treat and lead to increased mortality from otherwise treatable conditions. This underscores an urgent need for alternatives to conventional antibiotics.
Viruses that kill bacteria, known as phages, are potent antimicrobials that can be harnessed to treat infections. However, bacteria are equipped with a set of immune systems, including CRISPR-Cas, that protect against phage attack. These immune systems are problematic because they can reduce the efficacy of phage-based therapeutic treatments.
Asma Hatoum-Aslan, an associate professor of microbiology, studies the inner workings of bacterial immune systems with an emphasis on developing effective phage therapies. Her lab’s research centers around CRISPR-Cas and other immune systems in staphylococci, skin-dwelling bacteria that frequently cause antibiotic-resistant infections in humans. Two papers recently published by her lab describe the discovery of the first Type III-A anti-CRISPR protein and uncover a mechanism by which antiviral immune systems can spread and potentially compromise the efficacy of phage therapy.

Up close with a novel protein that inhibits CRISPR-Cas immunity
CRISPR-Cas immune systems use a special complex to detect and destroy nucleic acids with phage origins. This complex is composed of small RNAs bound to one or more CRISPR-associated Cas nucleases.
Of the six types of CRISPR-Cas systems, Type III are considered the most complex. While most CRISPR systems target either DNA or RNA invaders, Type III systems target both. Type III systems are also the only CRISPR variety known to utilize a second messenger signaling mechanism that stimulates Cas nucleases, and confers an added layer of protection by recruiting the help of housekeeping nucleases typically earmarked for other cellular activities. As a result, Type III CRISPR-Cas systems are incredibly effective in clearing phage infections.
This prompted the question: Have some phages evolved ways to fight back? To answer this, members of the Hatoum-Aslan lab screened a large collection of phages for the ability to evade Type III-A CRISPR-Cas systems and discovered a novel anti-CRISPR protein. Their findings, published in Nucleic Acids Research, highlight the ability of anti-CRISPR protein AcrIIIA1 to bind the CRISPR-associated complex and block its functions.
“Having access to a diverse collection of phages was key for this initial discovery,” Hatoum-Aslan said. “I developed a phage discovery course that I have been teaching since 2016. Each spring I have a classroom full of students looking for phages that infect Staphylococcus bacteria. So we’re sourcing from this large collection of phages that’s still growing.”
After identifying phages with anti-CRISPR activity, the researchers’ next challenge was determining which specific genes were responsible. After sifting through a pool of over 200 genes — many with unknown functions — the lab was successful in identifying acrIIIA1, the first Type III-A anti-CRISPR gene, in an effort Hatoum-Aslan coined “genetics gymnastics.”
By mating pairs of related phages that were resistant to the CRISPR system, lab members narrowed down their location of interest, zeroing in on a short segment of about 2,000 nucleotides. Cloning and testing multiple genes in this region allowed Hatoum-Aslan and her students to pinpoint the one ultimately responsible for anti-CRISPR activity.
Additional experiments revealed that AcrIIIA1 is unique in its composition; it is a small protein that binds strongly to small RNAs, including fragmented tRNAs, which are part of the cell’s protein-building machinery.
“We’re not entirely sure how those RNA fragments are helping the phage escape CRISPR, but we think they might be indirectly preventing housekeeping nucleases from degrading the phage’s genetic material through competitive inhibition,” Hatoum-Aslan said. “If you throw damaged RNA at these nucleases as a distraction, they’ll have other things to chew on. In the meantime, the phage gets to complete its replication and escape.”
Hatoum-Aslan hopes that phages engineered with anti-CRISPR proteins will be more effective in treating antibiotic-resistant infections when used in therapeutic applications.
“One of the benefits of teaching Phage Discovery is amassing this collection of phages that we can share with clinicians who are using phage therapy to resolve stubborn infections,” Hatoum-Aslan said. “We recently connected with an orthopedic surgeon in Pittsburgh and have been sending him some of our wild-caught S. epidermidis phages to treat patients with infections in their medical implants.”
The lab’s long-term goal is designing therapeutic phages that can overcome CRISPR-Cas and other defenses by equipping them with proteins like AcrIIIA1.
But while phage therapy has shown promise in some case studies, it has yet to become a routine treatment in the United States. One downstream issue concerning researchers is the sheer volume of anti-phage defenses employed by bacteria, and the potential for phage resistance mechanisms to spread rapidly. In a complementary paper, Hatoum-Aslan’s lab dug deeper into the antiviral arsenal in staphylococci.
Mobilizing defenses: SCCmec cassettes as a source of antiviral spread
Seeking to better understand the full battery of antiviral defenses in staphylococci, Hatoum-Aslan and her team identified all known defenses and their genomic locations in over 1,000 strains of S. aureus and S. epidermidis. Their analysis revealed a major pathway through which antiviral defenses can spread.
This analysis, described in Nature Communications, suggested that staphylococci carry many of their defenses in or near mobile segments of DNA known as SCCmec cassettes. Cassettes are discrete regions containing clusters of genes that can pop in or out of the genome. SCCmec cassettes can cut themselves out of a genome or paste themselves back in as a single discrete element. These cassettes can also escape their parent bacterium and transfer into a nearby, unrelated bacterium in a process called horizontal gene transfer.
“SCCmec casettes are notorious for spreading resistance to methicillin, which is one of our last-resort antibiotics against staph infections,” Hatoum-Aslan said. “Once we realized these cassettes and neighboring regions are hotspots for anti-phage defenses, we wanted to look deeper into their impact on defense movements.”
Members of the Hatoum-Aslan lab determined that in addition to cutting and pasting SCCmec cassettes, enzymes encoded within SCCmec cassettes can also cut and paste adjacent segments of DNA that carry multiple anti-phage immune systems. They also discovered that phage infection stimulates the release of these cassettes from cells, further facilitating their spread. These findings have implications for phage therapy, which must contend with increasing phage resistance.
“There is still a lot we don’t know about antiviral defenses in bacteria,” Hatoum-Aslan said. “It’s a completely wide-open field, but the bottleneck is figuring out how defense systems work. What is the trigger alerting the system to a phage infection? How does the system eliminate the phage while preventing damage to the cell?.”
Going forward, Hatoum-Aslan’s lab is working to identify and characterize new immune systems and how phages naturally adapt.
“Uncovering the details of this evolutionary process is very informative,” Hatoum-Aslan said. “It helps us design therapeutic phages that will be effective for many years to come.”